Linear Inference Sessions¶
An overview and example of a linear inference model for performing tasks such as GWAS in HFL.
The built-in model package iai_linear_inference trains a bundle of linear models for the target of interest against a specified list of predictors. It obtains the coefficients and variance estimates, and also calculates the p-values from the corresponding hypothesis tests. Linear inference is particularly useful for genome-wide association studies (GWAS), to identify genomic variants that are statistically associated with a risk for a disease or a particular trait.
This is a horizontal federated learning (HFL) model package.
Follow the instructions in the Installing the SDK section to prepare a local test environment for this tutorial.
Overview of the iai_linear_inference package¶
There are two strategies available in the package:
LogitRegInference- for use when the target of interest is binaryLinearRegInference- for use when the target is continuous
Example model_config for a binary target:
model_config_logit = {
"strategy": {"name": "LogitRegInference", "params": {}},
"seed": 23, # for reproducibility
}
Example model_config for a continuous target:
model_config_linear = {
"strategy": {"name": "LinearRegInference", "params": {}},
"seed": 23, # for reproducibility
}
Example data_config:
data_config_logit = {
"target": "y",
"shared_predictors": ["x1", "x2"],
"chunked_predictors": ["x0", "x3", "x10", "x11"]
}
The data_config dictionary should include the following three fields.
target: the target column of interestshared_predictors: predictor columns that should be included in all linear models. For example, the confounding factors like age, gender in GWAS.chunked_predictors: predictor columns that should be included in the linear model one at a time. For example, the gene expressions in GWAS.
Note: The columns in all the fields can be specified as either names/strings or indices/integers.
With this example data configuration, the session trains four logistic regression models with y as the target, and x1, x2 plus any one of x0, x3, x10, x11 as predictors.
Create a linear inference training session¶
Dataset information
Specify the datasets to be used in the session. These datasets must be registered in your workspace before they can be used in the notebook. Information about registering datasets is available here for AWS and here for Azure.
consumer_train_name = `demo_carrier_train`
consumer_test_name = `demo_carrier_test`
provider_train_name = `demo_provider_train`
provider_test_name = `demo_provider_test`
Set up the task builder
A task builder is an object that manages the individual tasks or clients that are involved in a session. This is generally boilerplate code - you only need to specify the task builder name. For example: iai_tb_aws_consumer.
from integrate_ai_sdk.taskgroup.taskbuilder.integrate_ai import IntegrateAiTaskBuilder
from integrate_ai_sdk.taskgroup.base import SessionTaskGroup
iai_tb_aws_consumer = IntegrateAiTaskBuilder(client=client,task_runner_id="")
iai_tb_aws_provider = IntegrateAiTaskBuilder(client=client,task_runner_id="")
Note: you do not need to specify the task runner name, but the variable name is required. This variable will be removed in an upcoming release.
Create a session
For this example, there are two (2) clients and the model is trained over five (5) rounds.
# Example training session
logit_session = client.create_fl_session(
name="Testing linear inference session",
description="I am testing linear inference session creation using a task runner through a notebook",
min_num_clients=2,
num_rounds=5,
package_name="iai_linear_inference",
model_config=model_config_logit,
data_config=data_config_logit
).start()
logit_session.id //prints the sesssion ID, for reference
name(str) - Name to set for the sessiondescription(str) - Description to set for the sessionmin_num_clients(int) - Number of clients required to connect before the session can beginnum_rounds(str) - Number of rounds of federated model training to performpackage_name(str) - Name of the model package to be used in the session. Useiai_linear_inferencefor linear inference sessions.model_config(dict) - Contains the model configuration to be used for the sessiondata_config(dict) - Contains the data configuration to be used for the session
Start the linear inference training session¶
This example demonstrates starting a training session with two tasks (clients).
#Create a task group
task_group_context = (
SessionTaskGroup(logit_session)
.add_task(iai_tb_aws_consumer.hfl(train_dataset_name=consumer_train_name, test_dataset_name=consumer_test_name))
.add_task(iai_tb_aws_provider.hfl(train_dataset_name=provider_train_name, test_dataset_name=provider_test_name))
.start()
)
Wait for the session to complete. You can view the session status on the Sessions page in your workspace, or by polling the SDK for session status.
View training metrics and model details¶
Once the session is complete, you can view the training metrics and model details such as the model coefficients and p-values. In this example, since there are a bundle of models being trained, the metrics are the average values of all the models.
training_session_logit.metrics().as_dict()
# Example output
{'session_id': '3cdf4be992',
'federated_metrics': [{'loss': 0.6931747794151306},
{'loss': 0.6766608953475952},
{'loss': 0.6766080856323242},
{'loss': 0.6766077876091003},
{'loss': 0.6766077876091003}],
'client_metrics': [{'user@integrate.ai:dedbb7e9be2046e3a49b28b0131c4b97': {'test_loss': 0.6931748060977674,
'test_accuracy': 0.4995,
'test_roc_auc': 0.5,
'test_num_examples': 4000},
'user@integrate.ai:339d50e453f244ed9cb2662ab2d3bb66': {'test_loss': 0.6931748060977674,
'test_accuracy': 0.4995,
'test_roc_auc': 0.5,
'test_num_examples': 4000}},
{'user@integrate.ai:dedbb7e9be2046e3a49b28b0131c4b97': {'test_num_examples': 4000,
'test_loss': 0.6766608866775886,
'test_roc_auc': 0.5996664746664747,
'test_accuracy': 0.57625},
'user@integrate.ai:339d50e453f244ed9cb2662ab2d3bb66': {'test_num_examples': 4000,
'test_loss': 0.6766608866775886,
'test_accuracy': 0.57625,
'test_roc_auc': 0.5996664746664747}},
{'user@integrate.ai:339d50e453f244ed9cb2662ab2d3bb66': {'test_loss': 0.6766080602706078,
'test_accuracy': 0.5761875,
'test_num_examples': 4000,
...
'user@integrate.ai:339d50e453f244ed9cb2662ab2d3bb66': {'test_accuracy': 0.5761875,
'test_roc_auc': 0.5996632246632246,
'test_num_examples': 4000,
'test_loss': 0.6766078165060236}}],
'latest_global_model_federated_loss': 0.6766077876091003}
Plot the metrics¶
training_session_logit.metrics().plot()
Example output:
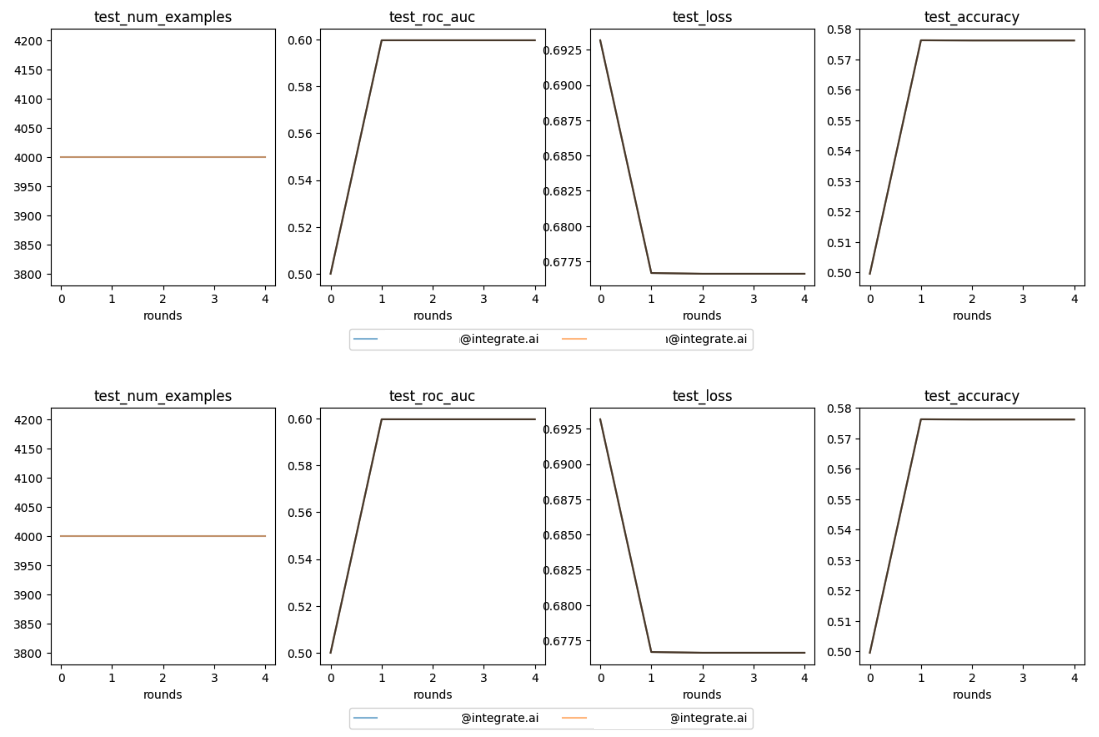
Retrieve the trained model¶
# Example of retrieving p-values
pv = model_logit.p_values()
pv
#Example p-value output:
x0 112.350396
x3 82.436540
x10 0.999893
x11 27.525280
dtype: float64
The LinearInferenceModel object can be retrieved using the model’s as_pytorch method. The relevant information, such as p-values, can be accessed directly from the model object.
model_logit = training_session_logit.model().as_pytorch()
Retrieve the p-values¶
The .p_values() function returns the p-values of the chunked predictors.
Summary information¶
The .summary method fetches the coefficient, standard error, and p-value of the model corresponding to the specified predictor.
#Example of fetching summary
summary_x0 = model_logit.summary("x0")
summary_x0`
Example summary output:
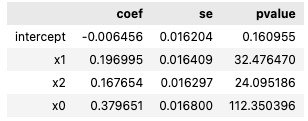
Making predictions from a linear inference session¶
from torch.utils.data import DataLoader
from integrate_ai_sdk.packages.LinearInference.dataset import ChunkedTabularDataset
ds = ChunkedTabularDataset(path=f"{data_path}/test.parquet", **data_config_logit)
dl = DataLoader(ds, batch_size=len(ds), shuffle=False)
x, _ = next(iter(dl))
y_pred = model_logit(x)
y_pred
You can also make predictions with the resulting bundle of models when the data is loaded by the ChunkedTabularDataset from the iai_linear_inference package. The predictions will be of shape (n_samples, n_chunked_predictors) where each column corresponds to one model from the bundle.
#Example prediction output:
tensor([[0.3801, 0.3548, 0.4598, 0.4809],
[0.4787, 0.3761, 0.4392, 0.3579],
[0.5151, 0.3497, 0.4837, 0.5054],
...,
[0.7062, 0.6533, 0.6516, 0.6717],
[0.3114, 0.3322, 0.4257, 0.4461],
[0.4358, 0.4912, 0.4897, 0.5110]], dtype=torch.float64)